- Patients Patients
Reproductive Genetics Testing
Patient Resources
Cost & Billing
- Providers Providers
- Genetic Counseling
- Login Login
- Estimate My Cost

MaterniT Redraw
Your patients deserve more, so you should expect more from an NIPS (NIPT)

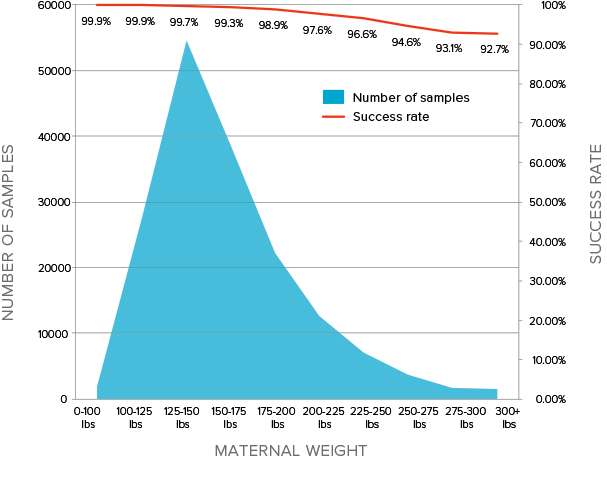
MaterniT 21 PLUS offers a very high success rate, especially in patients with higher maternal weight
A failed NIPS (NIPT) test result may lead to unnecessary patient redraws and/or diagnostic procedures, an issue often compounded by increasedmaternal weight. MaterniT 21 PLUS has a very high success rate inpatients with elevated maternal weight:
- 97.6% in patients that weigh between 200-225 lbs1
- 92.7% in patients that weigh 300+ lbs1
Some NIPSs (NIPTs) have a success rate as low as 72.5% (27.5%corresponding failure rate) in patients weighing over 200 lbs.2
97.6%
success rate at 200-225lbx
Highly reliable, extensively validated, the performance you should expect from the pioneer of NIPS (NIPT)
The MaterniT 21 PLUS test offers very low published and commercial non-reportable rates for trisomies 13, 18, and 21.

The MaterniT 21 PLUS test has been validated in clinical studies that tested samples from more than 2,100 pregnant women. The table below shows values for aneuploid samples in patients across four clinical studies, and accuracy for fetal sex within an additional publication.
| Positive results | Sensitivity | Specificity |
|---|---|---|
210 of 212 - trisomy 213,4 | 99.1% | 99.9% |
59 of 59 - trisomy 184 | > 99.9% | 99.6% |
11 of 12 - trisomy 134 | 91.7% | 99.7% |
8 of 8 multiple gestations: 7 of trisomy 21 | 1 of trisomy 138 | > 99.9% detection rate | |
Fetal sex9 | 99.4% accuracy | |
25 of 26 combined sex chromosome aneuploidies10 | 96.2% | 99.7% |
In a high-risk group, MaterniT 21 PLUS showed a positive predictive value greater than 97.9% for trisomy 21.11
References
- Wardrop J, McCullough R, Boomer T, et al. Maternal weight – impact on noninvasive prenatal testing (NIPT). Clinical poster presented at ACMG annual meeting, Florida 2016.
- Dar P, Curnow KJ, Gross SJ, et al. Clinical experience and follow-up with large scale single-nucleotide polymorphism-based noninvasive prenatal aneuploidy testing. Am J Obstet Gynecol. 2014 Nov; 211(5); 527.e1-527.e17.
- Palomaki GE, Deciu C, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet Med. 2011;13(11):913-920.
- Palomaki GE, Deciu C, Kloza EM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13, as well as Down syndrome: An international collaborative study. Genet Med. 2012;14(3):296-305.
- Norton ME, Brar H, Weiss J, et al. Non-invasive chromosomal evaluation (NICE) study: results of a multicenter, prospective, study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012 Aug;207(2):137.e1-8.
- Bianchi DW, Platt LD, Goldberg JD, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012 May;119(5):890-901.
- Pergament E, Cuckle H, Phil D, et al. Single-nucleotide polymorphism-based noninvasive prenatal screening in a high-risk and low-risk cohort. Obstet Gynecol. 2014 August; 124(20 1): 210–218.
- Canick JA, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to identify Down syndrome and other trisomies in multiple gestations. Prenat Diagn. 2012 Aug;32(8):730-4.
- Mazloom, A, Oeth P, Wang T, et al. Accuracy of noninvasive prenatal sex determination using massively parallel sequencing in samples from a large clinical validation study. Poster presented at American Society of Human Genetics annual meeting; 2012 Nov 6-10; San Francisco, CA.
- Mazloom AR, Džakula Ž, Oeth P. et al. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell-free DNA from maternal plasma. Prenat Diagn. 2013 Jun;33(6):591-7.
- Porreco RP, Garite TJ, Maurel K, et al. Noninvasive prenatal screening for fetal trisomies 21, 18, 13 and the common sex chromosome aneuploidies from maternal blood using massively parallel genomic sequencing of DNA. Am J Obstet Gynecol. 2014 Oct;211(4):365.e1-128.


